The polymerase chain reaction (PCR) is the one of the most powerful techniques that has been developed recently in the area of recombinant DNA research. PCR has had a major impact on many areas of molecular cloning and genetics. With this technique, a target sequence of DNA can be amplified a billion-fold in several hours. Amplification of particular segments of DNA by PCR is distinct from the amplification of DNA during cloning and propagation within a host cell. The procedure is carried out entirely in vitro. In addition to its use in many molecular cloning strategies, PCR is also used in the analysis of gene expression, forensic analysis where minute samples of DNA are isolated from a crime scene, and diagnostic tests for genetic diseases.
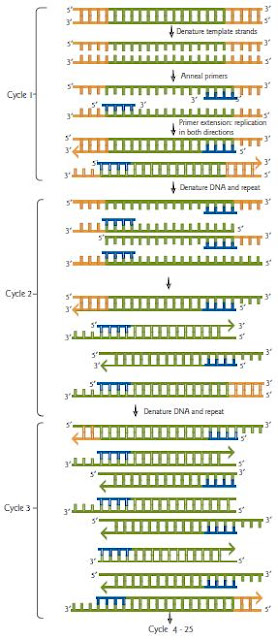
PCR is a DNA polymerase reaction. As with any DNA polymerase reaction it requires a DNA template and a free 3′-OH to get the polymerase started. The template is provided by the DNA sample to be amplified and the free 3′-OH groups are provided by site-specific oligonucleotide primers. The primers are complementary to each of the ends of the sequence that is to be amplified. Note that in vivo DNA polymerase would use an RNA primer, but a more stable, more easily synthesized DNA primer is used in vitro. The three steps of the reaction are denaturation, annealing of primers, and primer extension (Fig):
1 Denaturation. In the first step, the target sequence of DNA is heated to denature the template strands and
render the DNA single-stranded.
2 Annealing. The DNA is then cooled to allow the primers to anneal, that is, to bind the appropriate complementary strand. The temperature for this step varies depending on the size of the primer, the GC content, and its homology to
the target DNA. Primers are generally DNA oligonucleotides of approximately 20 bases each.
3 Primer extension. In the presence of Mg2+, DNA polymerase extends the primers on both strands from 5′ to 3′ by its polymerase activity. Primer extension is performed at a temperature optimal for the particular polymerase that is used.
Currently, the most popular enzyme for this step is Taq polymerase, the DNA polymerase from the thermophilic (heat-loving) bacteria Thermus aquaticus. This organism lives in hot springs that can be near boiling and thus requires a thermostable polymerase.
These three steps are repeated from 28 to 35 times. With each cycle, more and more fragments are generated with just the region between the primers amplified. These accumulate exponentially. The contribution of strands with extension beyond the target sequence becomes negligible since these accumulate in a linear manner. After 25 cycles in an automated thermocycler machine, there is a 225 amplification of the target sequence. PCR products can be visualized on a gel stained with nucleic acid-specific fluorescent compounds such as ethidium bromide or SYBR green. The error rate of Taq is 2 × 10−4. If an error occurs
early on in the cycles, it could become prominent.
Other polymerases, such as Pfu, have greater fidelity. Pfu DNA polymerase is from Pyrococcus furiosus. Base misinsertions that may occur infrequently during polymerization are rapidly excised by the 3′ → 5′ exonuclease (proofreading) activity of this enzyme.
When Kary Mullis first developed the PCR method in 1985, his experiments used E. coli DNA polymerase. Because E.coli DNA polymerase is heat-sensitive, its activity was destroyed during the denaturation step at 95°C. Therefore, a new aliquot of the enzyme had to be added in each cycle. The purification, and ultimately the cloning, of the DNA polymerase from T. aquaticus made the reaction much simpler. In his first experiments, Mullis had to move the reaction manually between the different temperatures. Fortunately, this procedure has been automated by the development of thermal cyclers. These instruments have the capability of rapidly switching between the different temperatures that are required for the PCR reaction. Thus the reactions can be set up and placed in the thermal cycler, and the researcher can return several hours later (or the next morning) to obtain the products.

No comments:
Post a Comment